Recently, the team led by Professor Chen Limeng, Director of the Department of Nephrology at PUMCH, in collaboration with Researcher Jiang Lan's team from the China National Center for Bioinformation and others, published a paper in Nature Methods (IF=36.1), a tier 1 journal among the top 5% as ranked by the Chinese Academy of Sciences. The paper introduced UDA-seq, their newly developed high-throughput single-cell multi-omics sequencing technology, and its successful application in frozen kidney biopsy tissues of ultralow amounts. Compared to traditional single-cell sequencing technologies, this solution is more efficient and convenient, providing an essential technical tool for the precision diagnosis and treatment of complex and rare kidney diseases. This research was supported by the National High-Level Hospital Clinical Research Funding, among others.
Kidney tissue contains more than 50 cell types. Single-cell sequencing technology can characterize cell-specific gene expression and regulatory features and describe disease-related cell subpopulations and intercellular interaction networks, potentially revealing pathogenesis and exploring new strategies for precision medicine. However, single-cell sequencing technology is expensive and requires large amounts of fresh tissue specimens, limiting its clinical applicability on a large scale.
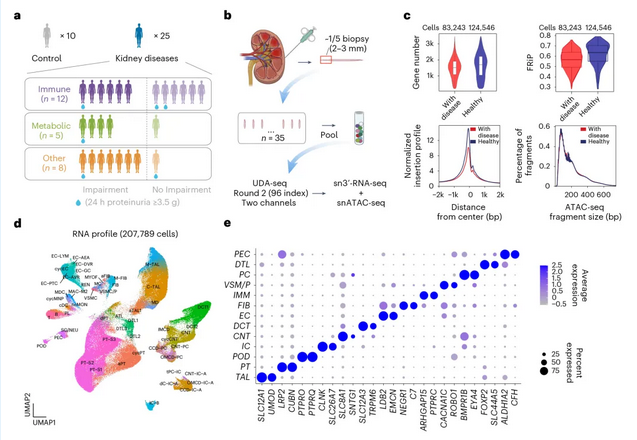
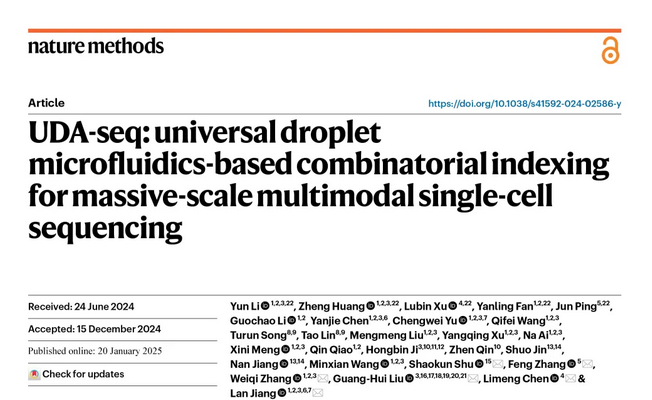
▲ UDA-seq was used for batch library construction and single-cell sequencing of 35 kidney biopsy tissues, obtaining dual-omics data from over 200,000 cells in a single experiment
UDA-seq, the new-generation high-throughput single-cell multi-omics sequencing technology, innovatively introduces a two-round barcode combinatorial indexing based on existing droplet microfluidics and applies genetic diversity information as natural barcodes, achieving high-throughput, low-cost dual-omics analysis of frozen biopsy tissues of ultralow amounts.
The research team used UDA-seq to sequence tiny clinical kidney biopsy tissues 2-3mm in length, completing the analysis of kidney specimens from 35 patients in a single experiment and collecting gene expression and chromatin accessibility information from over 200,000 cells. Based on this information, the team identified cell subpopulations associated with proteinuria and kidney function impairment caused by different etiologies and analyzed key transcription factors in their gene expression regulatory networks.
Director Chen Limeng explained that UDA-seq significantly reduces single-cell sequencing costs, enabling single-cell molecular pathology research and clinical applications using residual clinical specimens. Using this method to construct single-cell molecular atlases of kidney diseases can lay a foundation for analyzing disease pathogenesis and exploring therapeutic targets, further advancing precision medicine for complex and rare kidney diseases.
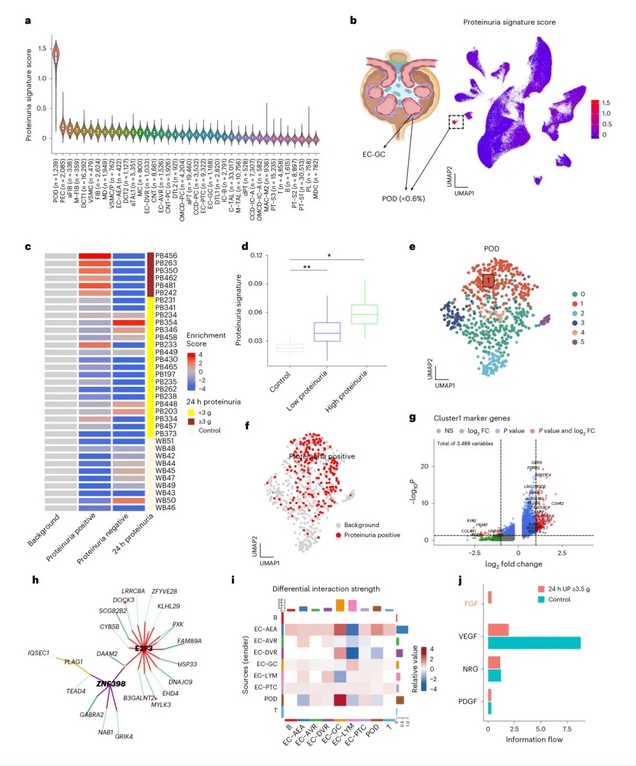
▲ UDA-seq single-cell dual-omics sequencing can be used to identify rare cell subpopulations associated with important clinical features and analyze key transcription factors in their gene regulatory networks
Written by Xu Lubin
Edited by Fu Tanping and Chen Xiao
