Recently, Researcher Yang Huayu from the Department of Liver Surgery and Chief Physician Kang Weiming from the Department of General Surgery at PUMCH published significant research findings in the internationally renowned journal Molecular Cancer (IF=33.9, a tier 1 journal ranked among the top 5% by the Chinese Academy of Sciences). The team innovatively constructed patient-derived 3D bioprinted gastric cancer models that can precisely mimic the histological characteristics, genomic profiles, and drug responses of gastric cancer tissues. This approach has the advantages of rapid model construction, low cost, and high fidelity. By moving beyond empirical chemotherapy selection, this model provides a reliable foundation for precision-guided treatment of gastric cancer. This research was supported by the National High-Level Hospital Clinical Research Funding and the National Natural Science Foundation of China.
Gastric cancer ranks as the fifth most prevalent malignancy globally and exhibits profound tumor heterogeneity. Traditional predictive approaches struggle to accurately estimate treatment efficacy, while existing patient-derived organoid and xenograft models have disadvantages such as prohibitive costs and inherent batch effect.
To overcome these challenges, the research team first needed to develop suitable "bioink". Through systematic screening of different hydrogels, the team identified a composite bioink that consisted of 6.25% gelatin methacryloyl (GelMA) and 0.5% hyaluronic acid methacryloyl (HAMA). The bioink not only possesses ideal mechanical properties but also perfectly mimics the extracellular matrix (ECM) environment in which gastric cancer cells survive. Subsequently, the team selected 33 tissue samples with adequate volume and no microbial contamination, and successfully produced their 3D printed gastric cancer (3DP-GC) models.
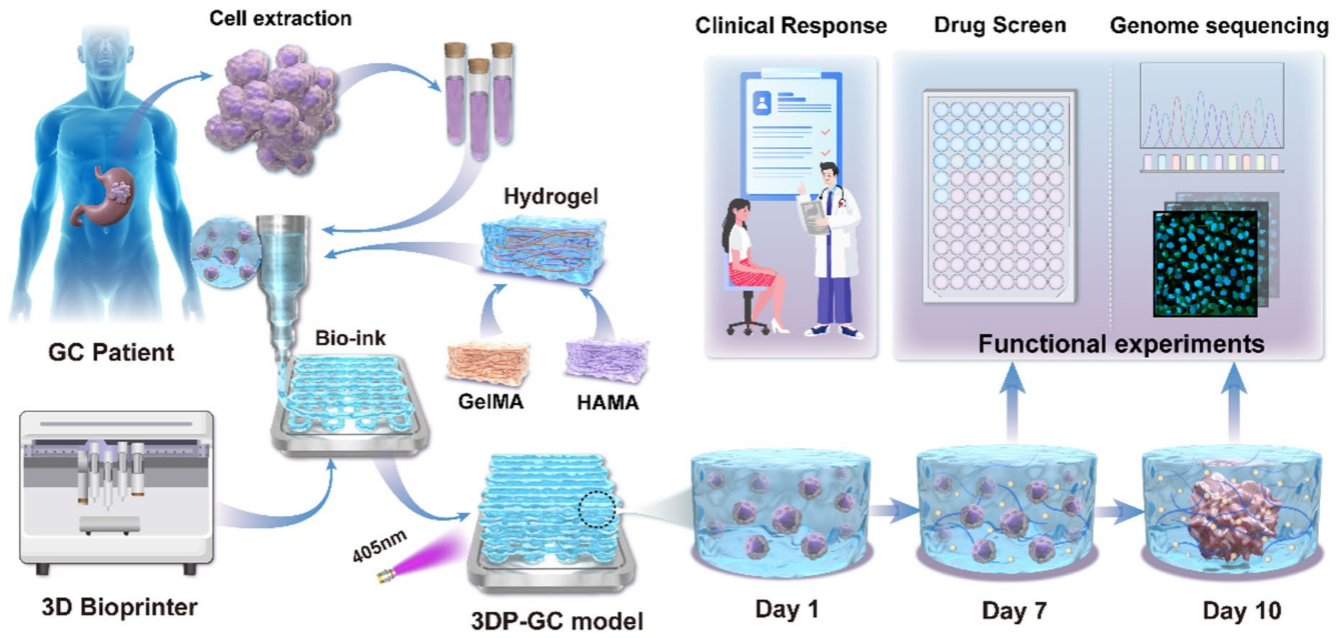
▲Schematic diagram of the fabrication process for patient-derived 3DP-GC models and their application in predicting clinical drug responses in patients
The model completely preserved key characteristics of the original tumor. Whole-exome sequencing revealed substantial concordance in single nucleotide variations (SNVs) between the 3DP-GC models and their respective original tumor tissues. The 3DP-GC models revealed a 100% mutation rate of the APC gene, consistent with the primary gastric cancers. Additionally, recurrent TP53, RNF43 and other mutations were consistently detected in both 3DP-GC models and parental tumors. The models thus demonstrated their potential to preserve the patient’s genomic characteristics.
The model demonstrated equally impressive performance in drug testing. Evaluation of the model’s response to first-line chemotherapy agents such as 5-fluorouracil and oxaliplatin showed that the model's drug sensitivity was significantly correlated with patients' clinical treatment outcomes, enabling discrimination between chemotherapy-sensitive and chemotherapy-resistant patients. The model also demonstrated potential for targeted therapy screening. Testing of HER2 and VEGFR2-positive models revealed that targeted drug efficacy was highly correlated with target expression levels, while small-molecule tyrosine kinase inhibitors demonstrated more pronounced concentration-dependent inhibitory effects, providing guidance for targeted therapy selection.
The 3DP-GC model has significant advantages. In contrast to traditional xenograft models, which require months to construct, the bioprinted model can be constructed in approximately one week. The combination of its low costs and readiness for batch production makes the model suitable for high-throughput drug screening while preserving stromal and immune cells within the tumor microenvironment. Follow-up analysis demonstrated that model predictions were highly concordant with clinical outcomes in 12 neoadjuvant chemotherapy patients and 5 adjuvant chemotherapy patients.
Yang Huayu stated that the team will continue to integrate multi-omics and artificial intelligence algorithms to optimize bioink formulations and drug combination predictions, with the goal of opening up new avenues for personalized treatment of malignant tumors.
Reviewed by Yang Huayu
